AI-Driven Drug Discovery Breakthrough: Turning the Tide on Antibiotic Resistance
In a major leap for medical science and biotechnology, researchers at the Massachusetts Institute of Technology (MIT) have announced a game-changing advancement in AI drug discovery and the fight against antibiotic resistance. Their study, published in Cell on August 14, 2025 (Cell, 2025), demonstrates that generative AI can design entirely novel antibiotic compounds — including two front-runners, NG1 and DN1 — that show potent activity against multidrug-resistant pathogens such as Neisseria gonorrhoeae (gonorrhea) and Staphylococcus aureus (MRSA) in preclinical testing.
1. Medical Breakthrough: MIT’s AI System Generates Millions of Antibiotic Compounds
The research team led by Professor James J. Collins at MIT’s IMES and the MIT Jameel Clinic used generative AI to explore vast chemical space in search of novel molecules.
Revolutionary Scale of Discovery
The team generated over 36 million candidate compounds using computational generative platforms (MIT News, 2025). The top compounds are structurally distinct from existing antibiotic classes — meaning they do not resemble known drugs and therefore may avoid existing resistance mechanisms (Cell, 2025).
Two Breakthrough Lead Molecules
Two lead molecules emerged: NG1, which showed compelling activity against N. gonorrhoeae (including resistant strains), and DN1, which demonstrated efficacy against MRSA in a mouse skin infection model (MIT News, 2025; Fierce Biotech, 2025).
Novel Mechanisms of Action
NG1 appears to disrupt the bacterial outer membrane via a novel target (LptA), rather than the usual cell wall or ribosome inhibitors (MIT Jameel Clinic, 2025).
From a clinical perspective, this signals a possible new antibiotic class — much needed as global innovation has stagnated. Over the past four decades, few truly novel classes have been introduced while resistance escalates.
2. Scientific Significance: NG1 and DN1 Against Resistant Strains
Effectiveness Against Key Drug-Resistant Pathogens
NG1 showed potent activity against drug-resistant N. gonorrhoeae — a Gram-negative bacterium with limited treatment options (MIT News, 2025). Reports indicate NG1 is selective for pathogenic N. gonorrhoeae in preclinical assays (MIT Jameel Clinic, 2025).
DN1 demonstrated efficacy against methicillin-resistant S. aureus (MRSA) in a mouse skin infection model and exhibited strong bactericidal activity against multidrug-resistant S. aureus in vitro (Fierce Biotech, 2025). Several AI-designed compounds showed antibacterial effects with low human-cell toxicity (Cell, 2025).
Novel Mechanism & Structure
NG1 likely targets LptA in outer-membrane synthesis — a pathway not exploited by most current antibiotics (MIT Jameel Clinic, 2025). DN1 appears to interfere with bacterial membranes more broadly (MIT News, 2025). Because these scaffolds are de novo, cross-resistance risks may be reduced (Cell, 2025).
Why This Matters for Resistance
A 2019 global analysis estimated antimicrobial resistance (AMR) caused ~1.27 million deaths and contributed to ~5 million deaths worldwide (The Lancet, 2022). Generative AI opens regions of chemical space previously inaccessible to conventional screening (Cell, 2025).
3. Technology Deep Dive: How Generative AI Explores Chemical Space
Traditional vs AI-Powered Discovery
Traditional discovery screens existing libraries (104–106 compounds) then optimizes leads. Generative AI designs novel molecules from scratch, exploring far larger chemical spaces (MIT News, 2025).
The computational demands of such AI-driven discovery are driving partnerships like OpenAI’s AWS cloud deal, as researchers require substantial compute for training molecular design models.
Generative Methods Used
- Fragment-based generation: Starting from a promising fragment (e.g., F1), models (VAE + genetic algorithms) iteratively built larger molecules (Cell, 2025).
- Unconstrained de novo: Models generated molecules without a starting fragment — effectively “from a single atom” upwards (Fierce Biotech, 2025).
At each step, candidates were scored by property predictors trained on antibacterial datasets and toxicity panels for activity, human-cell toxicity, synthetic feasibility, and novelty (Cell, 2025).
This also underscores the need for advanced hardware — with surging demand for specialized AI chips — and improvements in thermal management for high-performance compute that power design pipelines.
Integration with Pharma Pipelines
After in-silico generation and filtering, top molecules were synthesized. Of 24 compounds manufactured, 7 were active; 2 leads (NG1, DN1) advanced to in vivo mouse models (Fierce Biotech, 2025). Prioritizing synthetic feasibility helps bridge design and manufacturability.
Potential for Personalized Antibiotics
Because models can tailor scaffolds, future workflows could match agents to a patient’s resistant strain or microbiome profile. While MIT’s work targeted gonorrhea and MRSA, the platform could be adapted to narrow-spectrum, targeted therapies to minimize collateral microbiome effects.
Research Methodology Overview
- Platforms: Fragment-based (VAE/GA) and unconstrained de novo
- Fragment space: ~45M known fragments (Gram-negative target)
- De novo: >29M compounds (Gram-positive target)
- Filters: Activity predictors, cytotoxicity, liabilities, novelty, synthetic accessibility
- Synthesis & testing: 24 synthesized; 7 active; 2 leads to in vivo models
- Mechanism: NG1 → LptA (outer-membrane synthesis); DN1 → MRSA skin infection efficacy
Sources: Cell (2025); MIT News (2025).
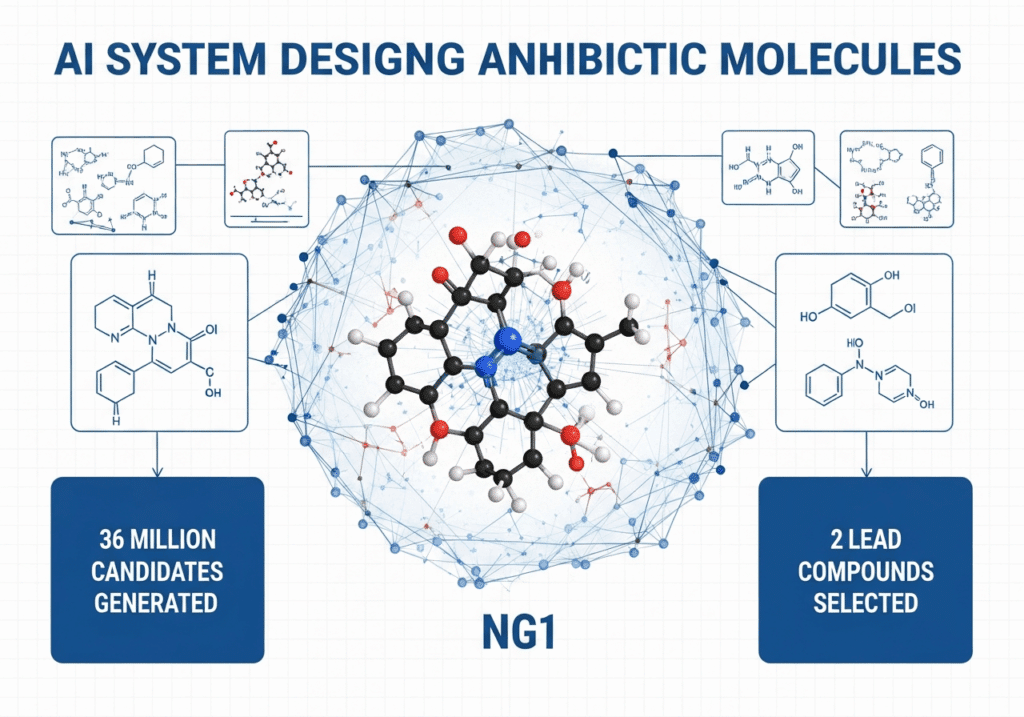
4. Clinical Timeline: From Laboratory to Patient Availability
These AI-designed antibiotics are preclinical. Human testing and regulatory steps will determine availability.
| Phase | Typical Duration | Key Activities | Projected Timeline* |
|---|---|---|---|
| Preclinical (in vitro, in vivo) | 1–2 years | Toxicology, PK, safety, animal infection models | Late preclinical (2025–2026) |
| Phase I (safety) | 1–2 years | Healthy volunteers, dose-finding | 2027–2028 |
| Phase II (efficacy/safety) | 1–3 years | Patient cohorts, dose selection | 2028–2030 |
| Phase III (confirmatory) | 2–4 years | Multicenter trials, endpoints | 2030–2032 |
| Approval & launch | ~1 year | FDA/EMA review, scale-up | 2032–2033 |
*Earliest potential availability ≈ 2032–2033 (optimistic). Timelines can extend due to toxicity, resistance, funding, or regulatory factors.
5. Industry Impact: R&D & Development Costs
Conventional antibiotic discovery can cost hundreds of millions and take years to find viable leads. Generative AI enables virtual screening/design at massive scale — reducing cost/time for hit-to-lead (MIT News, 2025). This yields more efficient pipelines, greater diversity, and a higher probability of discovering novel classes.
Although this study focuses on antibiotics, the tech is transferable (antivirals, antifungals, rare-disease small molecules, oncology). See recent pharma disruptions like oral weight-loss pill breakthroughs changing treatment paradigms.
Market & Investment Opportunities
Investors should track MIT’s Antibiotics-AI Project and organizations such as Phare Bio, which are advancing leads toward clinical application. The surge in AI stock valuations signals confidence in AI across sectors, but market volatility could affect biotech funding, underscoring the need for clear clinical progress and regulatory paths.
Pull incentives: subscription-based access (decoupling volume from revenue); AMR Action Fund/PASTEUR-style incentives for late-stage development; advanced market commitments for resistance-breaking agents.
Challenges & Caveats
Even with AI, bottlenecks remain: synthesis, manufacturability, regulatory risk, reimbursement (Fierce Biotech, 2025). Stewardship, short treatment courses, and low reimbursement complicate commercial viability. Integrating AI requires data-sharing, model validation, and alignment with wet-lab outcomes.
6. Global Health Implications
Antibiotic Resistance Today
The 2019 global analysis estimated AMR caused ~1.27M deaths and contributed to ~5M deaths worldwide (The Lancet, 2022). WHO lists AMR among the top global health threats. Priority pathogens include N. gonorrhoeae, MRSA, Acinetobacter baumannii, and Pseudomonas aeruginosa.
How the MIT Advance Helps
De novo, structurally distinct molecules open new killing mechanisms and may bypass entrenched resistance pathways (Cell, 2025). The specific activity of NG1 and DN1 against gonorrhea and MRSA highlights therapeutic potential.
Equity, Access & Diagnostics
For LMICs, access, affordability, and stewardship are critical. Pairing novel agents with rapid AST and sequencing will help preserve efficacy and guide use. Lower discovery costs via AI must be paired with policy frameworks, manufacturing capacity, and global distribution mechanisms.
Conclusion
MIT’s generative-AI antibiotic discovery marks a pivotal moment. The combination of structural novelty and early efficacy creates momentum toward revitalizing antimicrobial pipelines — but the journey from lab to patient still requires years of rigorous development.
Current status & next steps: Phare Bio is further modifying NG1/DN1 for additional preclinical testing. The team is extending platforms to other pathogens, including Mycobacterium tuberculosis and Pseudomonas aeruginosa.
Medical Disclaimer: This report summarizes preclinical research and is not medical advice. No AI-designed antibiotic discussed is approved for human use.
Sources & Citations
- Cell (Krishnan et al., 2025) – primary research
- MIT News (2025) – institutional coverage
- MIT Jameel Clinic / Community Jameel (2025) – mechanisms & lab notes
- Fierce Biotech (2025) – synthesis/testing details
- The Lancet (2022) – global AMR burden analysis
- World Health Organization – public health context